Get a free book sample
Fill in your information and the book will be sent to your email in no time

Bacterial Unknown Project. Original Photo by CDC on Unsplash
GIDEON’s Bacterial Unknowns Project allows microbiology professors to make and print dynamic dichotomous keys online. Using hundreds of decision trees, you can help your students identify over 2,000 pathogens with a few clicks of your mouse. That’s right. No more drawing elaborate dichotomous key flowcharts by hand or using a clunky word processing program.
GIDEON is one of the most well-known reference databases for infectious diseases and a trusted partner for clinicians and microbiologists worldwide. Together with microbiology professors, GIDEON designed an interactive decision tree tool to help identify bacteria, mycobacteria, and yeasts.
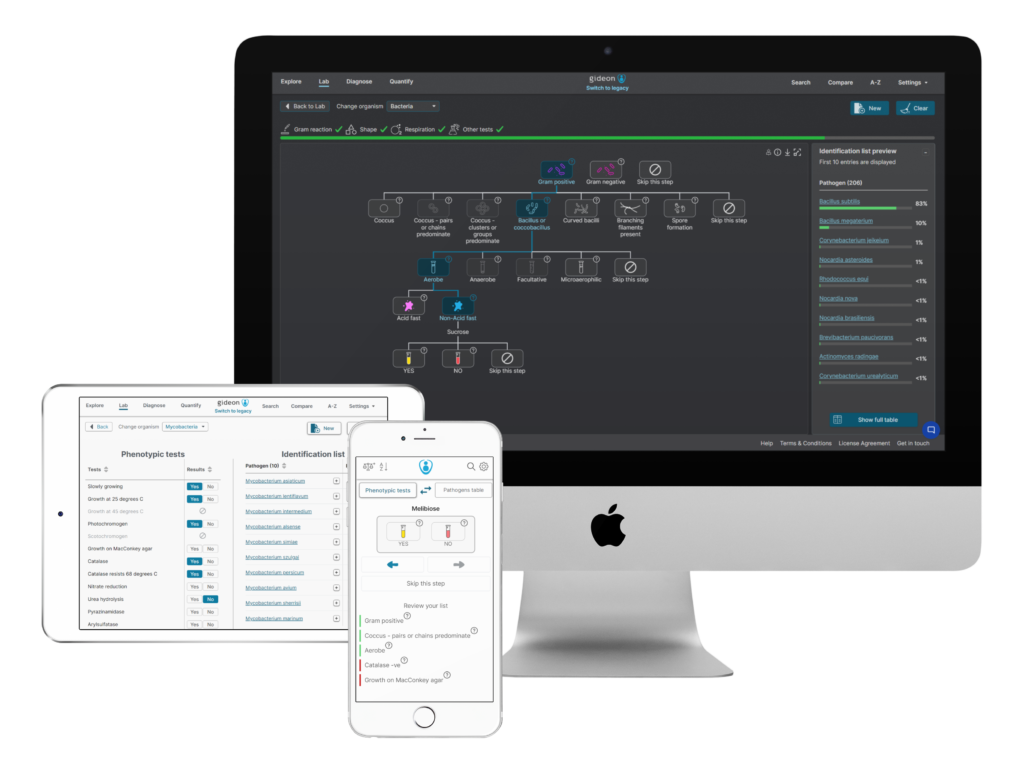
Sample screenshots of GIDEON Unknown Bacteria Project, decision tree.
A review from a satisfied customer:
“I was given 2 weeks to prepare the syllabus for two courses I needed to run in the fall semester, one in Public Health and the other in Pathogenic Microbiology. GIDEON was the perfect tool to build activities and learning around it made my job much easier!”
University of South Florida, Dr. Johnny El-Rady, Instructor (Microbiology and Genetics)
Professors agree that bacterial unknown projects are the ultimate test of a student’s understanding of pathogens in the classroom. Testing students with “mystery” bacteria allows them to combine their knowledge of pathogen molecular structures and biochemistry with essential laboratory techniques required to identify them.
However, these professors are also unanimous in accepting that preparing dichotomous keys for each class and semester can take time. As a result, students may attempt these active learning tests to identify microbes only once or twice in a semester.
Learning through the Unknown Bacteria Project is a great way for students to get ready for the real world. After all, professionals in the laboratory are counted upon to identify or confirm the presence of pathogens from an unknown organism. This expertise is essential whether in a hospital, clinic, or academic research lab. Working knowledge of fundamental laboratory techniques, conducting aseptic transfers, preparing media, and following a systematic flowchart to perform differential testing are invaluable skills.
For example, studying the science of pathogens for further research involves isolating and developing pure colonies to study. This step is critical to make sure the result of further testing is accurate, but new students may find this step difficult and need to practice. This is relevant even though labs are getting more automated than before. To be a professional writer, you have to know the basics of grammar and not rely entirely on auto-correct. Professional musicians need to understand how music notes work together in harmony, even if the software is available to help create music.
Knowing how to perform laboratory techniques correctly will help students identify pathogens in imperfect situations and troubleshoot when things go wrong. Bacterial Unknown Projects are also significant learning assignments because students learn to record and organize their data appropriately, work with their peers, and present their reasoning in written reports. These skills are priceless in a professional setting, private or academic, where we are often asked to defend or explain our work to other teams.
However, since 2020, the learning environment across the globe has changed considerably. Professors have been facing challenges when it comes to administering Unknown Bacteria Projects online.
The COVID-19 pandemic inserted a bit of a nonsense mutation in the lives of many microbiology professors. With so much uncertainty, lockdowns, and other restrictions, academic instruction went online. Didactic courses were easier to convert into an online format, but how do unknown projects translate to remote learning? After all, the very point of an unknown bacteria activity is to train students in a hands-on setting for wet lab techniques.
The Department of Molecular Biosciences at the University of Kansas at Lawrence published their experience in the Journal of Microbiology and Biology Education, March 2021. After administering a test to identify an unknown microbe for about 50 students, they conducted a survey and collected anonymous responses. 80% of students surveyed reported the online bacterial unknown project successful in increasing their skills. The project took over three weeks, with the last week to prepare for oral presentations.
Evaluations focused more on effective student collaborations, arriving at the correct result by asking for the right virtual “tests,” peer evaluations, and communicating their findings in an oral presentation instead of a written report. The entire project was done using images of stains. The paper discusses how delivering unknown projects online is not a substitute for wet lab experiences but a complementary teaching method.
Tighter budgets for lab equipment and resources means often having to test students on a limited range of bacteria for in-person wet-lab testing. Creating dynamic dichotomous keys online can help students identify pathogens that are important to learn about but may not be possible to administer in a teaching laboratory.
GIDEON’s decision trees for bacterial unknown projects are designed with busy microbiology professors in mind.
Create a dichotomous key in just 3 steps:
Step 1: Specify whether you want to identify a bacteria, mycobacteria, or yeast.
Step 2: If identifying bacteria, select: the gram reaction (gram-positive bacteria or gram-negative bacteria), bacterial shape (gram-negative rods, gram-positive rods, and more), respiration, and other lab tests (like negative urea test, blood work) by following the intuitive flowchart.
Step 3: Print your key, or export and share with your peers. It’s that simple.
The example below shows the flowchart for Bacillus Subtilis. On the right, you also get a list of the most probable bacteria based on your selections.
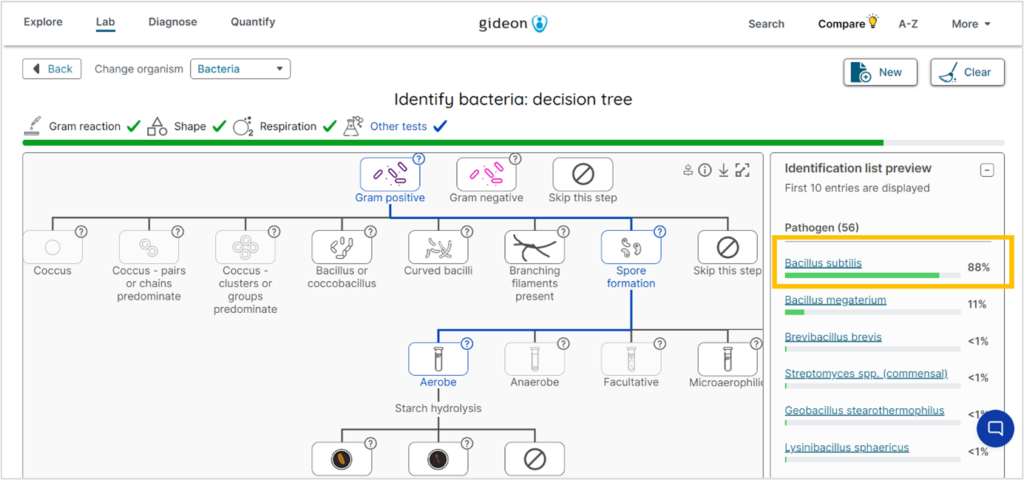
Dichotomous key for Bacillus Subtilis. Screenshot from GIDEON’s Bacterial Unknown Decision Tree.
Unknown Bacteria Project is considered to be one of the most effective ways to learn. Students apply their theoretical knowledge of molecular structure, biochemistry, and lab techniques in a practical wet lab setting. However, the process of preparing dichotomous keys for each test is cumbersome. Professors who are pressed for time find it challenging to draw decision trees by hand or use a word processing program.
Additionally, when the COVID-19 pandemic began, remote learning became the new normal. While adapting all courses to an online format can be hard, making Unknown Bacteria Projects applicable for remote learning is much trickier. Dynamic, online flowcharts and decision trees can help professors teach their students more effectively in online and wet laboratory settings.
GIDEON is one of the most well-known and comprehensive global databases for infectious diseases. Data is refreshed daily, and the GIDEON API allows medical professionals and researchers access to a continuous stream of data. Whether your research involves quantifying data, learning about specific microbes, or testing out differential diagnosis tools– GIDEON has you covered with a program that has met standards for accessibility excellence. You can also review our eBooks on Miscellaneous Intestinal Trematodes, Orbital and Eye Infections, Paragonimiasis, and more. Or check out our global status updates on countries like Rwanda, Saint Lucia, Antilles, and more!